The Species Are Related—The Question is How?
For thousands of years, people have looked at the life around them and tried to characterize all living species and to organize how they are related. One of the earliest-known theories was proposed by a man named Anaximander, who lived in ancient Greece around 600 BC. He proposed that humans must have emerged from some earlier, more primitive form of life. He reasoned that since human babies need to be cared for, humans must have come from an earlier, more capable animal.
About 150 years later, another scientist named Empedocles proposed that at one time in history, there had lived many strange and freakish creatures, in addition to those that populated the earth at that time, but that the strange and freakish “monsters” had not been able to survive. Empedocles could thus be credited with proposing a rudimentary form of the idea of natural selection.
Aristotle (384–322 BC) was one of the first scientists to propose a model to explain how the species could be related. (For example, could certain characteristics be transferred from one generation to the next?) He suggested that all living organisms—plants, animals, and humans—possess what he called “souls.” He went on to propose that all these organisms could be organized into a tree-like structure based on the type of soul each one has. He proposed that plants have a “vegetative” soul, possessing capabilities for growth, nutrition, and reproduction; that animals have a “sensitive” soul, possessing, in addition to the powers of the plant, the powers of perception, locomotion, and emotion; and that humans have the additional powers of thought and reason, i.e., the “rational” soul.
Proposed Mechanisms of Inheritance
Although many scientists have accepted that the species are related, they did not know how characteristics of parent organisms were transmitted to the next generation. Jean-Baptiste Lamarck (1744–1829) popularized the idea that a parent’s acquired characteristics could be transmitted to its offspring. For example, according to Lamarck’s proposal, giraffes have long necks because their parents stretched their necks to reach higher leaves. As offspring were born with slightly longer necks over generations, the necks of giraffes progressively grew longer. Lamarck postulated that there were little particles in the parents’ blood called “gemmules” that could transmit pieces of information to offspring through some unknown mechanism. With this perspective by which inheritance could be continuous, it was easy to believe that inheritance could also be slow and gradual.
About 100 years after Lamarck, Gregor Mendel (1822–1884), an Augustinian monk, experimented with peas to better understand inheritance. By carefully crossbreeding pea plants and tracking their characteristics through generations (yellow vs. green, wrinkled skin vs. smooth skin, etc.), he was able to show that inherited traits were, in fact, passed on in discrete “packets.” This clear finding stood in contrast to the concept of continuous inheritance, which would have been observed if the gemmule theory had been correct.
At about that same time, Alfred Russel Wallace (1823–1913) and Charles Darwin (1809–1882) were each developing their theories of inheritance and of how the species were related. They were friends but were also competitors in proposing and publishing the idea of evolution. Although Darwin won the race to publish with his now-infamous book, On the Origin of Species by Means of Natural Selection, or the Preservation of Favoured Races in the Struggle for Life, in 1859, he had sketched his idea of how he thought the species were related over 20 years earlier—long before he published his book.1
Not a Tree
While all these ideas contrasted with the biblical understanding of Genesis, which states that God created all the types of living creatures “according to their kinds,” there was no scientific evidence to support the Genesis account until the mid-twentieth century. At the time of Darwin, all a scientist could see with a microscope was the nucleus and sometimes the nucleolus. The cell appeared to be simple; no one had any idea about the complex molecular machinery inside. In fact, prior to 1970, an entire PhD thesis could be based on sequencing one gene. Nowadays, sequencing a gene can be done in less than a day, or even in a matter of hours.
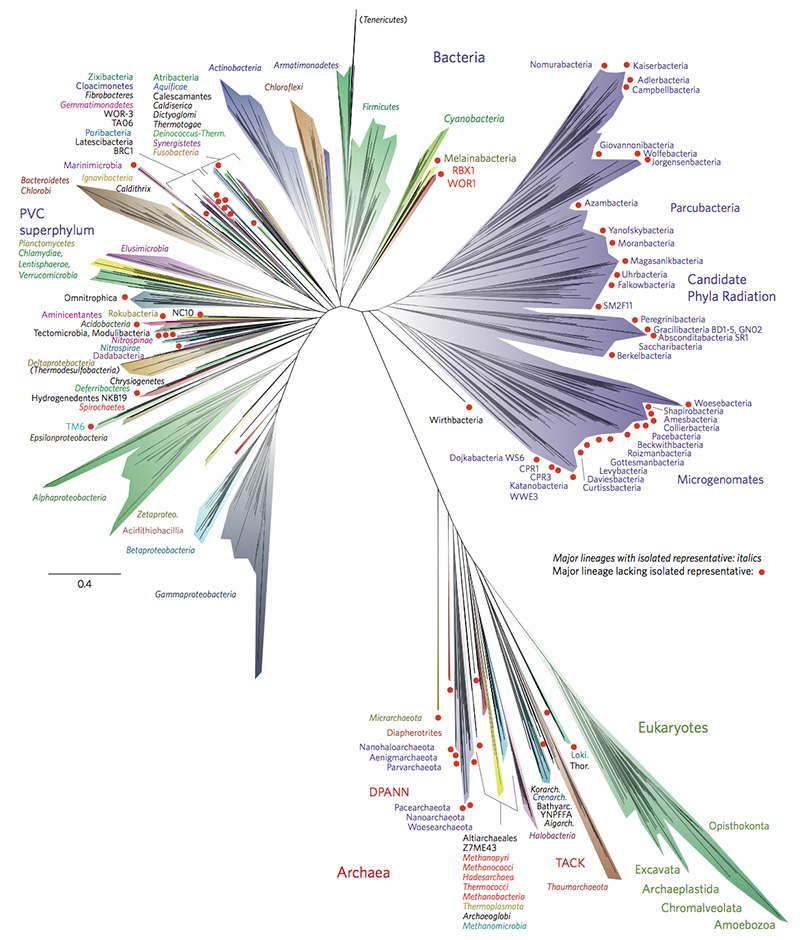
Whereas all that previous scientists had to go on was how the species looked, with the new knowledge that came in the wake of the DNA revolution, it became possible to sequence proteins, as well as two types of nucleic acids, DNA and mRNA. Research thus began on how best to draw an evolutionary tree based on genetic sequencing to explain how the species were related over time. But there were problems. Sequencing the DNA from a group of related species would result in a different tree than sequencing the mRNA from the same group of species. In fact, depending on which part of the genome was sequenced, or what type of nucleic acid was used, the sequencing produced different phylogenetic trees. (A phylogenetic tree depicts how the various species are related genetically. The length of each “branch” between two species indicates how closely or distantly they are related. See for example Figure 2.)
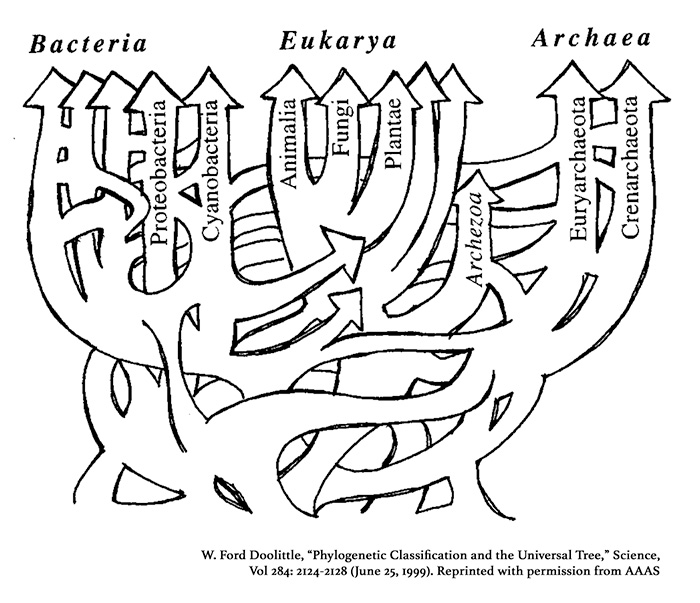
In 1999, W. Ford Doolittle reported that there was “evidence that most archaeal2 and bacterial genomes (and the inferred ancestral eukaryotic nuclear genome) contain genes from multiple sources” (see Figure 3).3 He characterized this phenomenon as “lateral gene transfer,” and he concludes that “no hierarchical universal classification can be taken as natural” and that “the history of life cannot properly be represented as a tree.”
Some scientists thought that if they could sequence even more genomes, they could sketch out a better clarification of how the species were related. In 2016, Hug et al. (there were 17 authors!) published the results of sequencing genomes from over 1000 organisms in the prestigious journal Nature.4 Their data showed a “dramatically expanded version of the tree of life.” One of the interesting and surprising findings they reported was that the Archaea (originally thought to be ancient life) was more closely related to the Eukarya (advanced life including plants, animals, and humans) than to the Bacteria domain.
Further research indicated that Doolittle could be right: the history of life cannot properly be represented as a tree. In a PNAS (Proceedings of the National Academy of Sciences) paper published in 2008, researchers reported that they had found a set of DNA transposons5 that are 96 percent identical across almost 2900 nucleotide bases in the genomes of rats, primates, bats, and tenrecs (from Madagascar); opossums (from South America); lizards (from Central and South America); and frogs (from West Africa).6 They fittingly named these transposons SPINs for SPACE INVADERS. The authors commented that, “Analysis of SPIN transposons across species reveals a combination of features that is incompatible with the traditional hypothesis of vertical inheritance” (emphasis added).7 They attributed these almost identical lengths of DNA to lateral gene transfer. But one wonders: how were these genes transmitted to organisms so widely spread around the world without being transmitted to more closely related species?
Another PNAS paper, published in 2019, reported that the echolocation ability in bats and whales was due to what the authors described as “convergent evolution.”8 Both the bat and the whale have 18 genes with substitutions allowing them to develop the ability to sense their surroundings through echolocation. One reporter for Stanford Medicine News Center commented, “Surprisingly, these very different species evolved their unique ability to use sound waves to navigate and identify obstacles and tasty morsels, be they mosquito or minnow, in part by acquiring identical mutations in their genomes—mutations not shared by other, more closely related species” (emphasis added).9
The above proposals for how the species are related genetically were loosely based on Darwin’s sketch of a tree-like relationship from 1837, and until recently, most proposed relationships were submitted by biologists. But, is the relationship really best represented by a tree?
Not By Chance
In 2018, a paper was published by Winston Ewert, a computer scientist who took a novel approach to modeling the relationship among the species. Rather than thinking in terms of a tree (reflecting the assumption of unguided descent with modification), Ewert asked if the relationship among the species could be better modeled by a dependency graph.10 A dependency graph depicts a technique used in software development by which programmers reuse modules of computer code (sometimes with minor modifications) across different software projects.
In other words, rather than using a tree-like, common-descent model, Ewert proposed a dependency-graph model which allows for tree-like relationships, but also incorporates a software developer’s structural reuse of code—a model suggestive of intentional design. He tested nine different biological databases and found that the common-descent model fit the data better than the null model, but that the dependency-graph model fit better than the common-descent model in all nine databases.
So, to return to the question at hand, how are the species related? The best explanation of Ewert’s finding is that there is some tree-like structure in modeling relationships among the species, but that there is also the additional design structure which could not happen by chance.
The fact that the relationship model approximates a tree-like structure is useful to research scientists who can, for example, experiment on yeast, nematodes, and fruit flies to understand the underlying biochemistry of human cellular biology. But the fact that there is also a design structure gives strong evidence of a designer.
Notes
1. Darwin’s Notebooks on Transmutation of Species Part I (1837).
2. Archaea are a classification of microorganisms that were thought to be the most primitive because their metabolism reflected the low-oxygen atmosphere in the hypothesized early earth.
3. W. Ford Doolittle, “Phylogenetic Classification and the Universal Tree,” Science Vol. 284 (June 25, 1999).
4. Hug et al., “A New View of the Tree of Life,” Nature Microbiology Vol. 11 (April 2016).
5. A transposon, sometimes called a “jumping gene,” is a piece of DNA that can move from one place in the genome to another, generally within the same genome.
6. Pace et al., “Repeated Horizontal Transfer of a DNA Transposon in Mammals and Other Tetrapods,” PNAS Vol. 105, No. 44 (Nov 4, 2008).
7. Gilbert et al., “Horizontal SPINning of Transposons,” Communicative & Integrative Biology Vol. 2 (Mar-Apr 2009).
8. Marcovitz et al., “A Functional Enrichment Test for Molecular Convergent Evolution Finds a Clear Protein-Coding Signal in Echolocating Bats and Whales,” PNAS Vol. 116, No. 42 (Sept 30, 2019).
9. Krista Conger, “Scientists Uncover Genetic Similarities Among Species that Use Sound to Navigate,” Stanford Medicine News Center (October 3, 2019).
10. Winston Ewert, “The Dependency Graph of Life,” BIO-Complexity Vol. 3 (2018).
is a (mostly) retired metallurgist. He received a BS in math and physics, before moving into materials science, then finishing with a Ph.D. in metallurgy. He is now spending some free time in exploring some of life’s generally accepted, but poorly supported ideas. For topics in biology, it helps a lot to have a wife who knows biology.
Get Salvo in your inbox! Elizabeth Siewertearned a BS in math and chemistry. She worked in the field of molecular biology at the University of Colorado for 15 years, carrying out research in genetics, where she learned a lot about biological systems. After her children were grown, she went on to earn a Ph.D. in biostatistics, thus combining her love for both math and biology. Her interest is in quantitatively modeling biological systems.
Get Salvo in your inbox! This article originally appeared in Salvo, Issue #71, Winter 2024 Copyright © 2026 Salvo | www.salvomag.com https://salvomag.com/article/salvo71/revisiting-the-tree-of-life


















